Note
Go to the end to download the full example code.
Simulation of a mixture model¶
Simulation of the mixture model, with estimation of the integration error.
Michel Bierlaire, EPFL Fri Jun 20 2025, 10:29:35
import sys
import numpy as np
from biogeme.biogeme import BIOGEME
from biogeme.expressions import Beta, Draws, MonteCarlo
from biogeme.models import logit
from biogeme.results_processing import EstimationResults
See the data processing script: Data preparation for Swissmetro.
from swissmetro_data import (
CAR_AV_SP,
CAR_CO_SCALED,
CAR_TT_SCALED,
CHOICE,
SM_AV,
SM_COST_SCALED,
SM_TT_SCALED,
TRAIN_AV_SP,
TRAIN_COST_SCALED,
TRAIN_TT_SCALED,
database,
)
try:
import matplotlib.pyplot as plt
PLOT = True
except ModuleNotFoundError:
print('Install matplotlib to see the distribution of integration errors.')
print('pip install matplotlib')
PLOT = False
Parameters to be estimated.
asc_car = Beta('asc_car', 0, None, None, 0)
asc_train = Beta('asc_train', 0, None, None, 0)
asc_sm = Beta('asc_sm', 0, None, None, 1)
b_cost = Beta('b_cost', 0, None, None, 0)
Define a random parameter, normally distributed, designed to be used for Monte-Carlo simulation.
b_time = Beta('b_time', 0, None, None, 0)
It is advised not to use 0 as starting value for the following parameter.
b_time_s = Beta('b_time_s', 1, None, None, 0)
b_time_rnd = b_time + b_time_s * Draws('b_time_rnd', 'NORMAL')
Definition of the utility functions.
v_train = asc_train + b_time_rnd * TRAIN_TT_SCALED + b_cost * TRAIN_COST_SCALED
v_swissmetro = asc_sm + b_time_rnd * SM_TT_SCALED + b_cost * SM_COST_SCALED
v_car = asc_car + b_time_rnd * CAR_TT_SCALED + b_cost * CAR_CO_SCALED
Associate utility functions with the numbering of alternatives.
v = {1: v_train, 2: v_swissmetro, 3: v_car}
Associate the availability conditions with the alternatives.
av = {1: TRAIN_AV_SP, 2: SM_AV, 3: CAR_AV_SP}
The estimation results are read from the pickle file.
try:
results = EstimationResults.from_yaml_file(
filename='saved_results/b05normal_mixture.yaml'
)
except FileNotFoundError:
print(
'Run first the script plot_b05normal_mixture.py in order to generate the '
'file b05normal_mixture.yaml.'
)
sys.exit()
Conditional to b_time_rnd, we have a logit model (called the kernel)
conditional_probability = logit(v, av, CHOICE)
We calculate the integration error. Note that this formula assumes independent draws, and is not valid for Halton or antithetic draws.
integral = MonteCarlo(conditional_probability)
integral_square = MonteCarlo(conditional_probability * conditional_probability)
variance = integral_square - integral * integral
error = (variance / 2.0) ** 0.5
And the value of the individual parameters.
numerator = MonteCarlo(b_time_rnd * conditional_probability)
denominator = integral
simulate = {
'Numerator': numerator,
'Denominator': denominator,
'Integral': integral,
'Integration error': error,
}
Create the Biogeme object.
biosim = BIOGEME(database, simulate, number_of_draws=10000)
biosim.model_name = 'b05normal_mixture_simul'
NUmber of draws
print(f'Number of draws: {biosim.number_of_draws}')
Number of draws: 10000
Simulate the requested quantities. The output is a Pandas data frame.
simulation_results = biosim.simulate(results.get_beta_values())
95% confidence interval on the log likelihood.
simulation_results['left'] = np.log(
simulation_results['Integral'] - 1.96 * simulation_results['Integration error']
)
simulation_results['right'] = np.log(
simulation_results['Integral'] + 1.96 * simulation_results['Integration error']
)
/Users/bierlair/python_envs/venv313/lib/python3.13/site-packages/pandas/core/arraylike.py:399: RuntimeWarning: invalid value encountered in log
result = getattr(ufunc, method)(*inputs, **kwargs)
print(f'Log likelihood: {np.log(simulation_results["Integral"]).sum()}')
Log likelihood: -5214.689681629532
print(
f'Integration error for {biosim.number_of_draws} draws: '
f'{simulation_results["Integration error"].sum()}'
)
Integration error for 10000 draws: 716.8094872624056
print(f'In average {simulation_results["Integration error"].mean()} per observation.')
In average 0.1059115672669039 per observation.
95% confidence interval
print(
f'95% confidence interval: [{simulation_results["left"].sum()} - '
f'{simulation_results["right"].sum()}]'
)
95% confidence interval: [-7583.2407468481015 - -2719.046943930449]
Post processing to obtain the individual parameters.
simulation_results['Beta'] = (
simulation_results['Numerator'] / simulation_results['Denominator']
)
Plot the histogram of individual parameters
if PLOT:
simulation_results['Beta'].plot(kind='hist', density=True, bins=20)
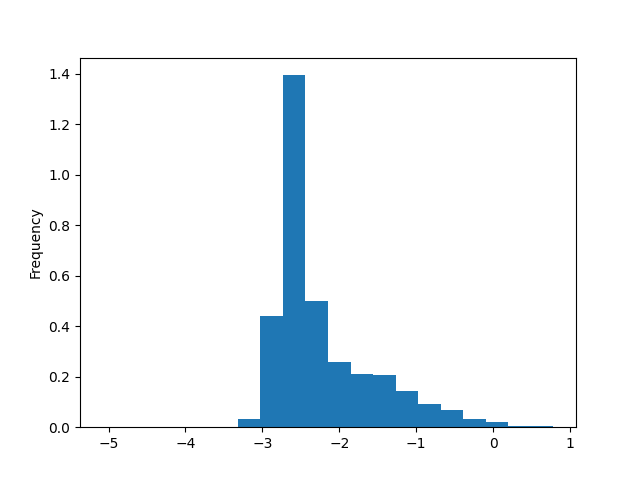
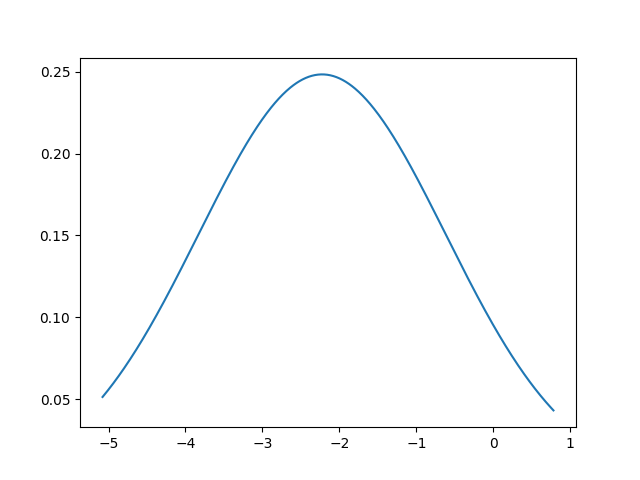
Plot the general distribution of Beta
def normalpdf(val: float, mu: float = 0.0, std: float = 1.0) -> float:
"""
Calculate the pdf of the normal distribution, for plotting purposes.
"""
d = -(val - mu) * (val - mu)
n = 2.0 * std * std
a = d / n
num = np.exp(a)
den = std * 2.506628275
p = num / den
return p
betas = results.get_beta_values(['b_time', 'b_time_s'])
x = np.arange(simulation_results['Beta'].min(), simulation_results['Beta'].max(), 0.01)
if PLOT:
plt.plot(x, normalpdf(x, betas['b_time'], betas['b_time_s']), '-')
plt.show()
Total running time of the script: (0 minutes 24.630 seconds)