Note
Go to the end to download the full example code.
Re-estimation of best models¶
After running the assisted specification algorithm for the 432 specifications in Combination of many specifications, we use post-processing to re-estimate all Pareto optimal models, and display some information about the algorithm. See Bierlaire and Ortelli (2023).
Michel Bierlaire, EPFL Sun Apr 27 2025, 18:38:57
from IPython.core.display_functions import display
from biogeme.biogeme import BIOGEME
from biogeme.results_processing import get_pandas_estimated_parameters
try:
import matplotlib.pyplot as plt
can_plot = True
except ModuleNotFoundError:
can_plot = False
import biogeme.biogeme_logging as blog
from biogeme.assisted import ParetoPostProcessing
from everything_spec import model_catalog, database
logger = blog.get_screen_logger(level=blog.INFO)
logger.info('Example b08selected_specification')
PARETO_FILE_NAME = 'saved_results/b07everything_assisted.pareto'
Example b08selected_specification
Create the biogeme object from the catalog.
the_biogeme = BIOGEME(database, model_catalog)
the_biogeme.model_name = 'b09post_processing'
Biogeme parameters read from biogeme.toml.
Create the post-processing object.
post_processing = ParetoPostProcessing(
biogeme_object=the_biogeme, pareto_file_name=PARETO_FILE_NAME
)
Pareto set initialized from file with 162 elements [13 Pareto] and 0 invalid elements.
Re-estimate the models.
all_results = post_processing.reestimate(recycle=True)
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000000
*** Initial values of the parameters are obtained from the file __b09post_processing_000000.iter
Cannot read file __b09post_processing_000000.iter. Statement is ignored.
Starting values for the algorithm: {}
As the model is not too complex, we activate the calculation of second derivatives. To change this behavior, modify the algorithm to "simple_bounds" in the TOML file.
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train_ref asc_train_diff_ b_time lambda_travel_t b_cost_train mu_existing asc_car_ref asc_car_diff_GA b_cost_car b_cost_swissmet Function Relgrad Radius Rho
0 -0.65 0.21 -1 1.4 -0.55 1.9 0.25 -0.26 0.14 -0.64 9.8e+03 0.23 1 0.43 +
1 -0.65 0.21 -1 1.4 -0.55 1.9 0.25 -0.26 0.14 -0.64 9.8e+03 0.23 0.5 -0.7 -
2 -0.26 0.4 -0.81 1.1 -0.4 1.8 -0.25 -0.38 -0.27 -0.46 8.6e+03 0.11 0.5 0.71 +
3 -0.42 0.67 -0.98 0.7 -0.64 2.3 -0.22 -0.51 -0.25 -0.52 8.2e+03 0.011 5 0.92 ++
4 -0.42 0.67 -0.98 0.7 -0.64 2.3 -0.22 -0.51 -0.25 -0.52 8.2e+03 0.011 0.64 -0.68 -
5 -0.24 1.1 -1.6 0.16 -0.81 1.6 -0.03 -0.34 -0.36 -0.64 8.2e+03 0.0074 0.64 0.75 +
6 -0.32 1 -1.6 0.35 -0.81 1.6 -0.083 -0.49 -0.34 -0.64 8.2e+03 0.0021 6.4 1 ++
7 -0.34 1 -1.5 0.36 -0.81 1.6 -0.09 -0.5 -0.34 -0.64 8.2e+03 1.6e-05 64 1 ++
8 -0.34 1 -1.5 0.36 -0.81 1.6 -0.09 -0.5 -0.34 -0.64 8.2e+03 1.6e-08 64 1 ++
Optimization algorithm has converged.
Relative gradient: 1.558341176595387e-08
Cause of termination: Relative gradient = 1.6e-08 <= 6.1e-06
Number of function evaluations: 24
Number of gradient evaluations: 15
Number of hessian evaluations: 7
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 9
Proportion of Hessian calculation: 7/7 = 100.0%
Optimization time: 0:00:04.278932
Calculate second derivatives and BHHH
File b09post_processing_000000.html has been generated.
File b09post_processing_000000.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000001
*** Initial values of the parameters are obtained from the file __b09post_processing_000001.iter
Cannot read file __b09post_processing_000001.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train b_time_ref b_time_diff_1st b_cost asc_car Function Relgrad Radius Rho
0 -0.75 -0.56 -0.47 -0.73 -0.27 8.7e+03 0.038 10 1.1 ++
1 -0.71 -0.88 -0.64 -0.84 -0.0093 8.6e+03 0.0053 1e+02 1.1 ++
2 -0.71 -0.92 -0.69 -0.87 0.0087 8.6e+03 0.00011 1e+03 1 ++
3 -0.71 -0.92 -0.69 -0.87 0.0087 8.6e+03 7.3e-08 1e+03 1 ++
Optimization algorithm has converged.
Relative gradient: 7.3355295711836e-08
Cause of termination: Relative gradient = 7.3e-08 <= 6.1e-06
Number of function evaluations: 13
Number of gradient evaluations: 9
Number of hessian evaluations: 4
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 4
Proportion of Hessian calculation: 4/4 = 100.0%
Optimization time: 0:00:00.417032
Calculate second derivatives and BHHH
File b09post_processing_000001.html has been generated.
File b09post_processing_000001.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000002
*** Initial values of the parameters are obtained from the file __b09post_processing_000002.iter
Cannot read file __b09post_processing_000002.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train_ref asc_train_diff_ b_time_ref b_time_diff_1st square_tt_coef cube_tt_coef b_cost_train b_cost_swissmet asc_car_ref asc_car_diff_GA b_cost_car Function Relgrad Radius Rho
0 0 0 0 0 0 0 0 0 0 0 0 1.1e+04 0.26 0.5 -1.5 -
1 0 0 0 0 0 0 0 0 0 0 0 1.1e+04 0.26 0.25 -0.051 -
2 -0.25 -0.00017 -0.25 -0.19 0 0 -0.25 0.25 0.0078 -0.023 -0.0062 9.4e+03 1.4 2.5 1 ++
3 -0.25 -0.00017 -0.25 -0.19 0 0 -0.25 0.25 0.0078 -0.023 -0.0062 9.4e+03 1.4 1.2 -5.6 -
4 -0.25 -0.00017 -0.25 -0.19 0 0 -0.25 0.25 0.0078 -0.023 -0.0062 9.4e+03 1.4 0.62 -3 -
5 -0.25 -0.00017 -0.25 -0.19 0 0 -0.25 0.25 0.0078 -0.023 -0.0062 9.4e+03 1.4 0.31 -1.4 -
6 -0.25 -0.00017 -0.25 -0.19 0 0 -0.25 0.25 0.0078 -0.023 -0.0062 9.4e+03 1.4 0.16 0.032 -
7 -0.36 0.019 -0.41 -0.26 0.15 -0.0031 -0.36 0.21 0.041 -0.052 0.022 9.2e+03 9.1 0.16 0.54 +
8 -0.36 0.019 -0.41 -0.26 0.15 -0.0031 -0.36 0.21 0.041 -0.052 0.022 9.2e+03 9.1 0.078 0.085 -
9 -0.38 0.037 -0.48 -0.27 0.12 0.0023 -0.38 0.13 0.059 -0.07 0.042 9.1e+03 5.4 0.078 0.16 +
10 -0.38 0.037 -0.48 -0.27 0.12 0.0023 -0.38 0.13 0.059 -0.07 0.042 9.1e+03 5.4 0.039 -4.2 -
11 -0.38 0.037 -0.48 -0.27 0.12 0.0023 -0.38 0.13 0.059 -0.07 0.042 9.1e+03 5.4 0.02 -2.8 -
12 -0.38 0.037 -0.48 -0.27 0.12 0.0023 -0.38 0.13 0.059 -0.07 0.042 9.1e+03 5.4 0.0098 -2 -
13 -0.38 0.037 -0.48 -0.27 0.12 0.0023 -0.38 0.13 0.059 -0.07 0.042 9.1e+03 5.4 0.0049 -1.1 -
14 -0.39 0.042 -0.48 -0.28 0.11 -0.0026 -0.39 0.13 0.061 -0.075 0.045 9e+03 11 0.0049 0.15 +
15 -0.39 0.043 -0.48 -0.28 0.11 0.0012 -0.39 0.12 0.061 -0.076 0.045 9e+03 5.3 0.0049 0.14 +
16 -0.39 0.043 -0.48 -0.28 0.11 0.0012 -0.39 0.12 0.061 -0.076 0.045 9e+03 5.3 0.0024 -0.6 -
17 -0.39 0.046 -0.49 -0.28 0.11 -0.0013 -0.39 0.12 0.064 -0.078 0.048 8.9e+03 4.2 0.0024 0.65 +
18 -0.39 0.047 -0.49 -0.28 0.11 -0.00081 -0.39 0.12 0.064 -0.079 0.048 8.9e+03 2.4 0.024 1.3 ++
19 -0.4 0.056 -0.5 -0.28 0.12 -0.00053 -0.4 0.093 0.064 -0.088 0.05 8.9e+03 5.5 0.024 0.63 +
20 -0.4 0.056 -0.5 -0.28 0.12 -0.00053 -0.4 0.093 0.064 -0.088 0.05 8.9e+03 5.5 0.012 -3.8 -
21 -0.4 0.056 -0.5 -0.28 0.12 -0.00053 -0.4 0.093 0.064 -0.088 0.05 8.9e+03 5.5 0.0061 -3.9 -
22 -0.4 0.056 -0.5 -0.28 0.12 -0.00053 -0.4 0.093 0.064 -0.088 0.05 8.9e+03 5.5 0.0031 -3.1 -
23 -0.4 0.056 -0.5 -0.28 0.12 -0.00053 -0.4 0.093 0.064 -0.088 0.05 8.9e+03 5.5 0.0015 -1.7 -
24 -0.4 0.056 -0.5 -0.28 0.12 -0.00053 -0.4 0.093 0.064 -0.088 0.05 8.9e+03 5.5 0.00076 -0.72 -
25 -0.4 0.056 -0.5 -0.28 0.12 -0.00053 -0.4 0.093 0.064 -0.088 0.05 8.9e+03 5.5 0.00038 -0.11 -
26 -0.4 0.056 -0.5 -0.29 0.12 -0.00091 -0.4 0.092 0.064 -0.088 0.05 8.9e+03 2.6 0.00038 0.32 +
27 -0.4 0.056 -0.5 -0.29 0.12 -0.00091 -0.4 0.092 0.064 -0.088 0.05 8.9e+03 2.6 0.00019 -0.56 -
28 -0.4 0.056 -0.5 -0.29 0.12 -0.00072 -0.4 0.092 0.063 -0.088 0.05 8.9e+03 1.9 0.00019 0.62 +
29 -0.4 0.056 -0.5 -0.29 0.12 -0.00075 -0.4 0.092 0.063 -0.088 0.05 8.9e+03 0.054 0.0019 1 ++
30 -0.4 0.057 -0.5 -0.29 0.12 -0.00076 -0.4 0.09 0.063 -0.089 0.05 8.9e+03 0.16 0.019 1 ++
31 -0.4 0.064 -0.51 -0.29 0.12 -0.00078 -0.41 0.071 0.063 -0.096 0.051 8.8e+03 0.051 0.19 1 ++
32 -0.44 0.14 -0.61 -0.33 0.18 -0.001 -0.49 -0.12 0.051 -0.17 0.052 8.7e+03 0.12 1.9 0.99 ++
33 -0.44 0.14 -0.61 -0.33 0.18 -0.001 -0.49 -0.12 0.051 -0.17 0.052 8.7e+03 0.12 0.95 -67 -
34 -0.44 0.14 -0.61 -0.33 0.18 -0.001 -0.49 -0.12 0.051 -0.17 0.052 8.7e+03 0.12 0.48 -3.4 -
35 -0.5 0.57 -0.82 -0.41 -0.011 -0.00022 -0.82 -0.6 -0.33 -0.5 -0.22 8.4e+03 0.32 0.48 0.89 +
36 -0.65 1 -1.2 -0.61 -0.022 -0.00017 -1.1 -0.85 -0.41 -0.72 -0.29 8.2e+03 0.15 4.8 1.1 ++
37 -0.65 1 -1.2 -0.61 -0.022 -0.00017 -1.1 -0.85 -0.41 -0.72 -0.29 8.2e+03 0.15 2.4 -3.2e+02 -
38 -0.65 1 -1.2 -0.61 -0.022 -0.00017 -1.1 -0.85 -0.41 -0.72 -0.29 8.2e+03 0.15 1.2 -1.1e+02 -
39 -0.65 1 -1.2 -0.61 -0.022 -0.00017 -1.1 -0.85 -0.41 -0.72 -0.29 8.2e+03 0.15 0.6 -22 -
40 -0.65 1 -1.2 -0.61 -0.022 -0.00017 -1.1 -0.85 -0.41 -0.72 -0.29 8.2e+03 0.15 0.3 -3.1 -
41 -0.65 1 -1.2 -0.61 -0.022 -0.00017 -1.1 -0.85 -0.41 -0.72 -0.29 8.2e+03 0.15 0.15 0.054 -
42 -0.77 1.1 -1.3 -0.69 -0.095 0.00018 -1.2 -0.78 -0.39 -0.75 -0.3 8.2e+03 8.8 0.15 0.74 +
43 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.15 0.59 +
44 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.075 -0.81 -
45 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.037 -0.61 -
46 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.019 -0.48 -
47 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.0093 -0.52 -
48 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.0047 -0.51 -
49 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.0023 -0.36 -
50 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.0012 -0.31 -
51 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.00058 -0.3 -
52 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.00029 -0.29 -
53 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 0.00015 -0.29 -
54 -0.69 1.2 -1.5 -0.75 -0.08 1.6e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.2e+03 14 7.3e-05 -0.29 -
55 -0.69 1.2 -1.5 -0.75 -0.079 8.9e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.1e+03 8.3 7.3e-05 0.62 +
56 -0.69 1.2 -1.5 -0.75 -0.08 7e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.1e+03 2 7.3e-05 0.81 +
57 -0.69 1.2 -1.5 -0.75 -0.08 7.4e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.1e+03 0.071 0.00073 0.98 ++
58 -0.69 1.2 -1.5 -0.75 -0.08 7.7e-05 -1.1 -0.87 -0.41 -0.8 -0.34 8.1e+03 0.015 0.0073 1 ++
59 -0.7 1.2 -1.5 -0.75 -0.086 0.0001 -1.2 -0.87 -0.4 -0.8 -0.34 8.1e+03 0.14 0.073 0.99 ++
60 -0.74 1.2 -1.5 -0.78 -0.096 0.00015 -1.2 -0.85 -0.38 -0.81 -0.33 8.1e+03 1.9 0.73 1 ++
61 -0.73 1.4 -1.8 -0.77 -0.11 0.00019 -1.1 -0.86 -0.31 -1 -0.33 8.1e+03 1.9 7.3 0.99 ++
62 -0.72 1.4 -1.8 -0.77 -0.1 0.00019 -1.1 -0.86 -0.3 -1 -0.32 8.1e+03 0.035 73 1 ++
63 -0.72 1.4 -1.8 -0.77 -0.1 0.00019 -1.1 -0.86 -0.3 -1 -0.32 8.1e+03 7.9e-05 7.3e+02 1 ++
64 -0.72 1.4 -1.8 -0.77 -0.1 0.00019 -1.1 -0.86 -0.3 -1 -0.32 8.1e+03 8e-08 7.3e+02 1 ++
Optimization algorithm has converged.
Relative gradient: 7.973438777139777e-08
Cause of termination: Relative gradient = 8e-08 <= 6.1e-06
Number of function evaluations: 122
Number of gradient evaluations: 57
Number of hessian evaluations: 28
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 65
Proportion of Hessian calculation: 28/28 = 100.0%
Optimization time: 0:00:00.985973
Calculate second derivatives and BHHH
File b09post_processing_000002.html has been generated.
File b09post_processing_000002.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000003
*** Initial values of the parameters are obtained from the file __b09post_processing_000003.iter
Cannot read file __b09post_processing_000003.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train_ref asc_train_diff_ asc_train_diff_ asc_train_diff_ b_time square_tt_coef cube_tt_coef b_cost_train mu_existing asc_car_ref asc_car_diff_GA asc_car_diff_on asc_car_diff_se b_cost_car b_cost_swissmet Function Relgrad Radius Rho
0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 1.1e+04 0.26 0.5 -2.7 -
1 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 1.1e+04 0.26 0.25 -0.39 -
2 -0.25 -0.0003 -0.25 -0.0098 -0.25 0 0 -0.25 1.2 0.013 -0.039 -0.011 -0.0031 -0.011 0.25 9.3e+03 2.1 2.5 1 ++
3 -0.25 -0.0003 -0.25 -0.0098 -0.25 0 0 -0.25 1.2 0.013 -0.039 -0.011 -0.0031 -0.011 0.25 9.3e+03 2.1 1.2 1 -
4 -0.25 -0.0003 -0.25 -0.0098 -0.25 0 0 -0.25 1.2 0.013 -0.039 -0.011 -0.0031 -0.011 0.25 9.3e+03 2.1 0.62 1 -
5 -0.25 -0.0003 -0.25 -0.0098 -0.25 0 0 -0.25 1.2 0.013 -0.039 -0.011 -0.0031 -0.011 0.25 9.3e+03 2.1 0.31 -1.7 -
6 -0.25 -0.0003 -0.25 -0.0098 -0.25 0 0 -0.25 1.2 0.013 -0.039 -0.011 -0.0031 -0.011 0.25 9.3e+03 2.1 0.16 -0.12 -
7 -0.32 0.037 -0.25 -0.011 -0.41 0.11 -0.0031 -0.35 1.3 0.047 -0.076 -0.018 -0.0056 0.017 0.18 9.2e+03 11 0.16 0.3 +
8 -0.32 0.037 -0.25 -0.011 -0.41 0.11 -0.0031 -0.35 1.3 0.047 -0.076 -0.018 -0.0056 0.017 0.18 9.2e+03 11 0.078 -0.51 -
9 -0.32 0.037 -0.25 -0.011 -0.41 0.11 -0.0031 -0.35 1.3 0.047 -0.076 -0.018 -0.0056 0.017 0.18 9.2e+03 11 0.039 -0.42 -
10 -0.32 0.037 -0.25 -0.011 -0.41 0.11 -0.0031 -0.35 1.3 0.047 -0.076 -0.018 -0.0056 0.017 0.18 9.2e+03 11 0.02 -0.36 -
11 -0.32 0.037 -0.25 -0.011 -0.41 0.11 -0.0031 -0.35 1.3 0.047 -0.076 -0.018 -0.0056 0.017 0.18 9.2e+03 11 0.0098 -0.34 -
12 -0.32 0.037 -0.25 -0.011 -0.41 0.11 -0.0031 -0.35 1.3 0.047 -0.076 -0.018 -0.0056 0.017 0.18 9.2e+03 11 0.0049 -0.32 -
13 -0.32 0.042 -0.24 -0.01 -0.4 0.11 0.0018 -0.35 1.3 0.045 -0.081 -0.023 -0.0096 0.02 0.17 9.1e+03 5.3 0.0049 0.17 +
14 -0.32 0.042 -0.24 -0.01 -0.4 0.11 0.0018 -0.35 1.3 0.045 -0.081 -0.023 -0.0096 0.02 0.17 9.1e+03 5.3 0.0024 -0.18 -
15 -0.32 0.044 -0.24 -0.0097 -0.4 0.11 -0.00066 -0.35 1.3 0.046 -0.084 -0.026 -0.012 0.021 0.17 9e+03 2.7 0.024 0.99 ++
16 -0.32 0.055 -0.23 -0.0095 -0.42 0.12 -0.00076 -0.36 1.3 0.045 -0.093 -0.032 -0.013 0.021 0.14 8.9e+03 0.55 0.24 1 ++
17 -0.3 0.17 -0.12 -0.0078 -0.57 0.22 -0.0011 -0.44 1.4 0.036 -0.18 -0.097 -0.02 0.015 -0.1 8.6e+03 0.52 2.4 0.97 ++
18 -0.3 0.17 -0.12 -0.0078 -0.57 0.22 -0.0011 -0.44 1.4 0.036 -0.18 -0.097 -0.02 0.015 -0.1 8.6e+03 0.52 1.2 0.97 -
19 -0.3 0.17 -0.12 -0.0078 -0.57 0.22 -0.0011 -0.44 1.4 0.036 -0.18 -0.097 -0.02 0.015 -0.1 8.6e+03 0.52 0.61 -33 -
20 -0.43 0.72 0.27 0.013 -0.93 -0.11 0.0002 -0.93 2 -0.17 -0.58 -0.31 -0.077 -0.18 -0.64 8.5e+03 7.3 0.61 0.3 +
21 -0.47 0.59 0.16 0.029 -1.5 -0.034 -0.00011 -0.63 1.9 -0.18 -0.48 -0.051 -0.11 -0.31 -0.68 8.2e+03 7.8 0.61 0.68 +
22 -0.47 0.59 0.16 0.029 -1.5 -0.034 -0.00011 -0.63 1.9 -0.18 -0.48 -0.051 -0.11 -0.31 -0.68 8.2e+03 7.8 0.31 -5.7 -
23 -0.47 0.59 0.16 0.029 -1.5 -0.034 -0.00011 -0.63 1.9 -0.18 -0.48 -0.051 -0.11 -0.31 -0.68 8.2e+03 7.8 0.15 -2.8 -
24 -0.47 0.59 0.16 0.029 -1.5 -0.034 -0.00011 -0.63 1.9 -0.18 -0.48 -0.051 -0.11 -0.31 -0.68 8.2e+03 7.8 0.076 -0.71 -
25 -0.49 0.6 0.16 0.03 -1.6 -0.11 0.00021 -0.66 2 -0.17 -0.48 -0.05 -0.11 -0.3 -0.65 8.1e+03 19 0.076 0.65 +
26 -0.49 0.67 0.23 0.039 -1.6 -0.1 0.00017 -0.72 2 -0.15 -0.48 -0.06 -0.12 -0.26 -0.59 8.1e+03 0.71 0.76 1 ++
27 -0.6 0.92 0.34 0.19 -2 -0.11 0.00019 -0.81 1.5 -0.16 -0.45 -0.057 -0.31 -0.27 -0.68 8.1e+03 1 0.76 0.87 +
28 -0.59 0.9 0.35 0.22 -2 -0.11 0.0002 -0.79 1.6 -0.17 -0.46 -0.063 -0.36 -0.24 -0.67 8.1e+03 0.27 7.6 1 ++
29 -0.59 0.9 0.35 0.22 -2 -0.11 0.0002 -0.78 1.6 -0.17 -0.45 -0.063 -0.37 -0.24 -0.66 8.1e+03 0.0069 76 1 ++
30 -0.59 0.9 0.35 0.21 -2 -0.11 0.0002 -0.78 1.6 -0.17 -0.45 -0.063 -0.37 -0.24 -0.66 8.1e+03 0.081 7.6e+02 1 ++
31 -0.59 0.9 0.35 0.21 -2 -0.11 0.0002 -0.78 1.6 -0.17 -0.45 -0.063 -0.37 -0.24 -0.66 8.1e+03 0.0051 7.6e+03 1 ++
32 -0.59 0.9 0.35 0.21 -2 -0.11 0.0002 -0.78 1.6 -0.17 -0.45 -0.063 -0.38 -0.24 -0.66 8.1e+03 0.00066 7.6e+04 1 ++
33 -0.59 0.9 0.35 0.21 -2 -0.11 0.0002 -0.78 1.6 -0.17 -0.45 -0.063 -0.38 -0.24 -0.66 8.1e+03 5.8e-06 7.6e+04 1 ++
Optimization algorithm has converged.
Relative gradient: 5.817355202609906e-06
Cause of termination: Relative gradient = 5.8e-06 <= 6.1e-06
Number of function evaluations: 69
Number of gradient evaluations: 35
Number of hessian evaluations: 17
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 34
Proportion of Hessian calculation: 17/17 = 100.0%
Optimization time: 0:00:03.824482
Calculate second derivatives and BHHH
File b09post_processing_000003.html has been generated.
File b09post_processing_000003.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000004
*** Initial values of the parameters are obtained from the file __b09post_processing_000004.iter
Cannot read file __b09post_processing_000004.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train b_time b_cost asc_car Function Relgrad Radius Rho
0 -0.76 -0.77 -0.7 -0.29 8.8e+03 0.04 10 1.1 ++
1 -0.66 -1.2 -0.77 -0.0015 8.7e+03 0.0064 1e+02 1.1 ++
2 -0.65 -1.3 -0.79 0.016 8.7e+03 0.00012 1e+03 1 ++
3 -0.65 -1.3 -0.79 0.016 8.7e+03 4e-08 1e+03 1 ++
Optimization algorithm has converged.
Relative gradient: 3.954408093567457e-08
Cause of termination: Relative gradient = 4e-08 <= 6.1e-06
Number of function evaluations: 13
Number of gradient evaluations: 9
Number of hessian evaluations: 4
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 4
Proportion of Hessian calculation: 4/4 = 100.0%
Optimization time: 0:00:00.333908
Calculate second derivatives and BHHH
File b09post_processing_000004.html has been generated.
File b09post_processing_000004.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000005
*** Initial values of the parameters are obtained from the file __b09post_processing_000005.iter
Cannot read file __b09post_processing_000005.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. Function Relgrad Radius Rho
0 1.1e+04 0.4 0.5 -0.56 -
1 1e+04 1.5 0.5 0.2 +
2 1e+04 1.5 0.25 0.2 -
3 1e+04 1.5 0.12 0.2 -
4 1e+04 1.5 0.062 -4.2 -
5 9.9e+03 7.1 0.062 0.48 +
6 9.9e+03 7.1 0.031 0.0017 -
7 9.6e+03 1.2 0.031 0.12 +
8 9.6e+03 1.2 0.016 -0.32 -
9 9.3e+03 0.58 0.16 0.92 ++
10 9.3e+03 0.58 0.078 0.92 -
11 9.3e+03 0.58 0.039 -15 -
12 9.3e+03 0.58 0.02 -16 -
13 9.3e+03 0.58 0.0098 -6.7 -
14 9.3e+03 3.5 0.0098 0.56 +
15 9.3e+03 0.11 0.0098 0.88 +
16 9.2e+03 0.44 0.098 0.97 ++
17 9.1e+03 0.41 0.98 1 ++
18 9.1e+03 0.41 0.49 -0.77 -
19 8.7e+03 10 0.49 0.45 +
20 8.7e+03 10 0.24 0.45 -
21 8.7e+03 10 0.12 0.45 -
22 8.7e+03 10 0.061 0.45 -
23 8.7e+03 10 0.031 0.45 -
24 8.7e+03 10 0.015 0.45 -
25 8.7e+03 10 0.0076 0.45 -
26 8.7e+03 10 0.0038 -1.7 -
27 8.7e+03 10 0.0019 -1.1 -
28 8.7e+03 10 0.00095 -0.63 -
29 8.7e+03 10 0.00048 -0.36 -
30 8.7e+03 10 0.00024 -0.065 -
31 8.7e+03 6.6 0.00024 0.51 +
32 8.7e+03 7.8 0.00024 0.3 +
33 8.7e+03 1.4 0.00024 0.85 +
34 8.7e+03 0.19 0.0024 1 ++
35 8.7e+03 1.2 0.024 1 ++
36 8.6e+03 0.16 0.24 1 ++
37 8.3e+03 0.89 0.24 0.62 +
38 8.2e+03 0.22 2.4 0.98 ++
39 8.1e+03 0.091 24 1.1 ++
40 8.1e+03 0.77 24 0.75 +
41 8e+03 0.5 2.4e+02 1.1 ++
42 8e+03 0.5 1.2e+02 1.1 -
43 8e+03 0.5 60 1.1 -
44 8e+03 0.5 30 1.1 -
45 8e+03 0.5 15 1.1 -
46 8e+03 0.5 7.5 1.1 -
47 8e+03 0.5 3.7 1.1 -
48 8e+03 0.5 1.9 -4.9e+02 -
49 8e+03 0.5 0.93 -2e+02 -
50 8e+03 0.5 0.47 -19 -
51 8e+03 0.5 0.23 -1.2 -
52 8e+03 2.4 0.23 0.72 +
53 8e+03 3.2 2.3 1 ++
54 8e+03 3.2 0.39 -3.9 -
55 8e+03 2.4 0.39 0.47 +
56 8e+03 8.4 3.9 0.94 ++
57 8e+03 0.1 39 1 ++
58 8e+03 0.00059 3.9e+02 1 ++
59 8e+03 9.3e-06 3.9e+03 1 ++
60 8e+03 3.1e-06 3.9e+03 1 ++
Optimization algorithm has converged.
Relative gradient: 3.120917028152968e-06
Cause of termination: Relative gradient = 3.1e-06 <= 6.1e-06
Number of function evaluations: 118
Number of gradient evaluations: 57
Number of hessian evaluations: 28
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 61
Proportion of Hessian calculation: 28/28 = 100.0%
Optimization time: 0:00:07.476546
Calculate second derivatives and BHHH
File b09post_processing_000005.html has been generated.
File b09post_processing_000005.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000006
*** Initial values of the parameters are obtained from the file __b09post_processing_000006.iter
Cannot read file __b09post_processing_000006.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train_ref asc_train_diff_ b_time_ref b_time_diff_1st b_cost asc_car_ref asc_car_diff_GA Function Relgrad Radius Rho
0 -0.93 1 -0.56 -0.4 -0.66 -0.29 -0.4 8.5e+03 0.041 10 1.1 ++
1 -1.1 1.4 -0.84 -0.56 -0.75 -0.01 -1.1 8.3e+03 0.0076 1e+02 1.1 ++
2 -1.2 1.5 -0.87 -0.62 -0.78 0.0051 -1.2 8.3e+03 0.00034 1e+03 1 ++
3 -1.2 1.5 -0.87 -0.62 -0.78 0.0051 -1.2 8.3e+03 6.4e-07 1e+03 1 ++
Optimization algorithm has converged.
Relative gradient: 6.381936270565892e-07
Cause of termination: Relative gradient = 6.4e-07 <= 6.1e-06
Number of function evaluations: 13
Number of gradient evaluations: 9
Number of hessian evaluations: 4
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 4
Proportion of Hessian calculation: 4/4 = 100.0%
Optimization time: 0:00:00.493179
Calculate second derivatives and BHHH
File b09post_processing_000006.html has been generated.
File b09post_processing_000006.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000007
*** Initial values of the parameters are obtained from the file __b09post_processing_000007.iter
Cannot read file __b09post_processing_000007.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train_ref asc_train_diff_ b_time_ref b_time_diff_com square_tt_coef cube_tt_coef b_cost asc_car_ref asc_car_diff_GA Function Relgrad Radius Rho
0 0 0 0 0 0 0 0 0 0 1.1e+04 0.26 0.5 -0.12 -
1 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 5 1.1 ++
2 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 2.5 -4.7 -
3 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 1.2 -4.3 -
4 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.62 -3.9 -
5 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.31 -3.6 -
6 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.16 -3.5 -
7 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.078 -3.7 -
8 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.039 -4.1 -
9 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.02 -4.3 -
10 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.0098 -3.1 -
11 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.0049 -2.4 -
12 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.0024 -2 -
13 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.0012 -1.5 -
14 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.00061 -0.87 -
15 -0.5 -0.0018 -0.5 -0.5 0 0 -0.41 0.082 -0.24 9.1e+03 9.7 0.00031 -0.074 -
16 -0.5 -0.0015 -0.5 -0.5 0.00031 -0.00031 -0.41 0.081 -0.24 9.1e+03 6.1 0.00031 0.69 +
17 -0.5 -0.0015 -0.5 -0.5 0.00031 -0.00031 -0.41 0.081 -0.24 9.1e+03 6.1 0.00015 -1.3 -
18 -0.5 -0.0015 -0.5 -0.5 0.00031 -0.00031 -0.41 0.081 -0.24 9.1e+03 6.1 7.6e-05 -0.74 -
19 -0.5 -0.0014 -0.5 -0.5 0.00038 -0.00023 -0.41 0.081 -0.24 9.1e+03 5.6 7.6e-05 0.4 +
20 -0.5 -0.0014 -0.5 -0.5 0.00046 -0.00026 -0.41 0.081 -0.24 9.1e+03 1.3 7.6e-05 0.84 +
21 -0.5 -0.0014 -0.5 -0.5 0.00053 -0.00025 -0.41 0.081 -0.24 9.1e+03 0.14 0.00076 0.99 ++
22 -0.5 -0.0013 -0.5 -0.5 0.0013 -0.00026 -0.41 0.081 -0.24 9.1e+03 0.35 0.0076 1 ++
23 -0.5 -0.00013 -0.51 -0.5 0.0089 -0.00029 -0.41 0.079 -0.24 9.1e+03 0.14 0.076 1 ++
24 -0.54 0.012 -0.57 -0.51 0.085 -0.0006 -0.42 0.064 -0.24 8.9e+03 0.53 0.76 0.98 ++
25 -0.72 0.77 -0.95 -0.3 0.13 -0.00078 -0.82 0.03 -0.55 8.4e+03 9.1 7.6 1 ++
26 -0.72 0.77 -0.95 -0.3 0.13 -0.00078 -0.82 0.03 -0.55 8.4e+03 9.1 3.8 -1.2e+02 -
27 -0.72 0.77 -0.95 -0.3 0.13 -0.00078 -0.82 0.03 -0.55 8.4e+03 9.1 1.9 -1e+02 -
28 -0.72 0.77 -0.95 -0.3 0.13 -0.00078 -0.82 0.03 -0.55 8.4e+03 9.1 0.95 -70 -
29 -0.72 0.77 -0.95 -0.3 0.13 -0.00078 -0.82 0.03 -0.55 8.4e+03 9.1 0.48 -15 -
30 -0.72 0.77 -0.95 -0.3 0.13 -0.00078 -0.82 0.03 -0.55 8.4e+03 9.1 0.24 -2.2 -
31 -0.96 0.95 -1.2 -0.29 -0.096 0.00012 -0.7 -0.0011 -0.62 8.4e+03 17 0.24 0.43 +
32 -1 1.1 -1.4 -0.34 -0.073 6.6e-05 -0.68 -0.066 -0.66 8.3e+03 10 2.4 0.94 ++
33 -1 1.1 -1.4 -0.34 -0.073 6.6e-05 -0.68 -0.066 -0.66 8.3e+03 10 0.62 -4 -
34 -0.93 1.7 -1.9 -0.54 -0.11 0.00021 -0.76 0.11 -0.87 8.2e+03 38 0.62 0.71 +
35 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.62 0.62 +
36 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.087 -2.1 -
37 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.044 -1.9 -
38 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.022 -2 -
39 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.011 -1.9 -
40 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.0055 -1.8 -
41 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.0027 -1.8 -
42 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.0014 -1.8 -
43 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.00068 -1.7 -
44 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.00034 -1.7 -
45 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 0.00017 -1.6 -
46 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 8.5e-05 -1.6 -
47 -0.82 1.6 -2.1 -1.2 -0.1 0.00022 -0.69 0.21 -1.2 8.2e+03 27 4.3e-05 -1.2 -
48 -0.82 1.6 -2.1 -1.2 -0.1 0.00018 -0.69 0.21 -1.2 8.2e+03 26 4.3e-05 0.55 +
49 -0.82 1.6 -2.1 -1.2 -0.1 0.00019 -0.69 0.21 -1.2 8.2e+03 0.64 0.00043 0.95 ++
50 -0.82 1.6 -2.1 -1.2 -0.1 0.00019 -0.69 0.21 -1.2 8.2e+03 0.033 0.0043 1 ++
51 -0.83 1.6 -2.1 -1.2 -0.11 0.00021 -0.7 0.21 -1.2 8.2e+03 0.55 0.043 0.99 ++
52 -0.87 1.6 -2.1 -1.2 -0.11 0.00021 -0.72 0.18 -1.2 8.2e+03 0.091 0.43 1 ++
53 -0.9 1.6 -2 -1.2 -0.11 0.0002 -0.71 0.17 -1.3 8.2e+03 0.14 4.3 1 ++
54 -0.9 1.6 -2 -1.2 -0.11 0.0002 -0.71 0.17 -1.3 8.2e+03 0.021 43 1 ++
55 -0.9 1.6 -2 -1.2 -0.11 0.0002 -0.71 0.17 -1.3 8.2e+03 0.0096 4.3e+02 1 ++
56 -0.9 1.6 -2 -1.2 -0.11 0.0002 -0.71 0.17 -1.3 8.2e+03 0.0092 4.3e+03 1 ++
57 -0.9 1.6 -2 -1.2 -0.11 0.0002 -0.71 0.17 -1.3 8.2e+03 3.9e-07 4.3e+03 1 ++
Optimization algorithm has converged.
Relative gradient: 3.897850964533427e-07
Cause of termination: Relative gradient = 3.9e-07 <= 6.1e-06
Number of function evaluations: 105
Number of gradient evaluations: 47
Number of hessian evaluations: 23
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 58
Proportion of Hessian calculation: 23/23 = 100.0%
Optimization time: 0:00:02.862128
Calculate second derivatives and BHHH
File b09post_processing_000007.html has been generated.
File b09post_processing_000007.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000008
*** Initial values of the parameters are obtained from the file __b09post_processing_000008.iter
Cannot read file __b09post_processing_000008.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train b_time b_cost_train b_cost_swissmet asc_car b_cost_car Function Relgrad Radius Rho
0 -0.59 -0.75 -0.92 -0.69 -0.42 -0.57 8.7e+03 0.053 10 1.1 ++
1 -0.19 -1.2 -1.5 -0.78 -0.39 -0.39 8.5e+03 0.021 1e+02 1.2 ++
2 -0.064 -1.3 -1.9 -0.82 -0.41 -0.38 8.4e+03 0.0039 1e+03 1.1 ++
3 -0.046 -1.3 -1.9 -0.82 -0.42 -0.38 8.4e+03 0.00012 1e+04 1 ++
4 -0.046 -1.3 -1.9 -0.82 -0.42 -0.38 8.4e+03 1e-07 1e+04 1 ++
Optimization algorithm has converged.
Relative gradient: 1.0424282105589559e-07
Cause of termination: Relative gradient = 1e-07 <= 6.1e-06
Number of function evaluations: 16
Number of gradient evaluations: 11
Number of hessian evaluations: 5
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 5
Proportion of Hessian calculation: 5/5 = 100.0%
Optimization time: 0:00:00.340770
Calculate second derivatives and BHHH
File b09post_processing_000008.html has been generated.
File b09post_processing_000008.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000009
*** Initial values of the parameters are obtained from the file __b09post_processing_000009.iter
Cannot read file __b09post_processing_000009.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. asc_train_ref asc_train_diff_ b_time_ref b_time_diff_com square_tt_coef cube_tt_coef b_cost_train mu_existing asc_car_ref asc_car_diff_GA b_cost_car b_cost_swissmet Function Relgrad Radius Rho
0 0 0 0 0 0 0 0 1 0 0 0 0 1.1e+04 0.26 0.5 -2.9 -
1 0 0 0 0 0 0 0 1 0 0 0 0 1.1e+04 0.26 0.25 -0.22 -
2 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 2.5 1 ++
3 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 1.2 1 -
4 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.62 1 -
5 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.31 1 -
6 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.16 1 -
7 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.078 1 -
8 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.039 -3 -
9 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.02 -3.4 -
10 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.0098 -3.7 -
11 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.0049 -4.2 -
12 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.0024 -2.3 -
13 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.0012 -1.6 -
14 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.00061 -0.94 -
15 -0.25 -0.0082 -0.25 -0.25 0 0 -0.25 1.2 0.25 -0.25 -0.025 0.25 9.2e+03 5.6 0.00031 -0.17 -
16 -0.25 -0.0079 -0.25 -0.25 0.00031 -0.00031 -0.25 1.3 0.25 -0.25 -0.025 0.25 9.2e+03 3.7 0.00031 0.63 +
17 -0.25 -0.0079 -0.25 -0.25 0.00051 -0.00021 -0.25 1.3 0.25 -0.25 -0.025 0.25 9.2e+03 2.9 0.00031 0.56 +
18 -0.25 -0.0078 -0.25 -0.25 0.00072 -0.00026 -0.25 1.3 0.25 -0.25 -0.025 0.25 9.2e+03 0.6 0.0031 0.92 ++
19 -0.25 -0.0073 -0.25 -0.25 0.0028 -0.00026 -0.25 1.3 0.25 -0.25 -0.026 0.25 9.2e+03 0.1 0.031 1 ++
20 -0.27 -0.0022 -0.28 -0.25 0.024 -0.00035 -0.27 1.3 0.24 -0.26 -0.03 0.25 9.1e+03 0.5 0.31 1 ++
21 -0.4 0.058 -0.59 -0.29 0.23 -0.0012 -0.43 1.4 0.2 -0.33 -0.07 0.19 8.9e+03 1.9 0.31 0.55 +
22 -0.29 0.17 -0.59 -0.26 0.1 -0.00065 -0.42 1.5 0.13 -0.42 -0.097 -0.11 8.5e+03 0.3 3.1 0.9 ++
23 -0.54 1 -0.98 -0.48 -0.13 0.00028 -0.68 1.8 -0.26 -0.43 -0.27 -0.56 8.3e+03 26 3.1 0.74 +
24 -0.54 1 -0.98 -0.48 -0.13 0.00028 -0.68 1.8 -0.26 -0.43 -0.27 -0.56 8.3e+03 26 0.88 -2.2 -
25 -0.54 1 -0.98 -0.48 -0.13 0.00028 -0.68 1.8 -0.26 -0.43 -0.27 -0.56 8.3e+03 26 0.44 -0.096 -
26 -0.6 0.94 -1.4 -0.57 -0.062 2.6e-05 -0.64 1.9 -0.26 -0.41 -0.3 -0.58 8.2e+03 27 0.44 0.55 +
27 -0.6 0.94 -1.4 -0.57 -0.062 2.6e-05 -0.64 1.9 -0.26 -0.41 -0.3 -0.58 8.2e+03 27 0.22 -3.7 -
28 -0.6 0.94 -1.4 -0.57 -0.062 2.6e-05 -0.64 1.9 -0.26 -0.41 -0.3 -0.58 8.2e+03 27 0.11 -1.6 -
29 -0.6 0.94 -1.4 -0.57 -0.062 2.6e-05 -0.64 1.9 -0.26 -0.41 -0.3 -0.58 8.2e+03 27 0.055 -0.51 -
30 -0.59 0.94 -1.4 -0.57 -0.12 0.00023 -0.64 1.9 -0.26 -0.41 -0.29 -0.59 8.1e+03 43 0.055 0.39 +
31 -0.53 0.96 -1.4 -0.58 -0.092 0.00014 -0.62 1.9 -0.26 -0.41 -0.28 -0.63 8.1e+03 14 0.055 0.88 +
32 -0.53 0.97 -1.5 -0.59 -0.1 0.00017 -0.65 1.9 -0.25 -0.41 -0.27 -0.6 8.1e+03 3.2 0.55 0.96 ++
33 -0.37 1 -1.9 -0.91 -0.11 0.0002 -0.86 1.5 -0.16 -0.53 -0.31 -0.69 8.1e+03 3.7 5.5 0.91 ++
34 -0.36 1 -1.9 -0.93 -0.11 0.0002 -0.82 1.5 -0.17 -0.56 -0.27 -0.67 8.1e+03 0.76 55 1 ++
35 -0.35 1 -1.9 -0.93 -0.11 0.0002 -0.82 1.6 -0.17 -0.56 -0.27 -0.66 8.1e+03 0.019 5.5e+02 1 ++
36 -0.35 1 -1.9 -0.93 -0.11 0.0002 -0.82 1.6 -0.17 -0.55 -0.27 -0.66 8.1e+03 1.8e-05 5.5e+03 1 ++
37 -0.35 1 -1.9 -0.93 -0.11 0.0002 -0.82 1.6 -0.17 -0.55 -0.27 -0.66 8.1e+03 8.4e-05 5.5e+04 1 ++
38 -0.35 1 -1.9 -0.93 -0.11 0.0002 -0.82 1.6 -0.17 -0.55 -0.27 -0.66 8.1e+03 9.9e-07 5.5e+04 1 ++
Optimization algorithm has converged.
Relative gradient: 9.9074111913857e-07
Cause of termination: Relative gradient = 9.9e-07 <= 6.1e-06
Number of function evaluations: 78
Number of gradient evaluations: 39
Number of hessian evaluations: 19
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 39
Proportion of Hessian calculation: 19/19 = 100.0%
Optimization time: 0:00:03.075296
Calculate second derivatives and BHHH
File b09post_processing_000009.html has been generated.
File b09post_processing_000009.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000010
*** Initial values of the parameters are obtained from the file __b09post_processing_000010.iter
Cannot read file __b09post_processing_000010.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. Function Relgrad Radius Rho
0 8.6e+03 0.055 1 0.68 +
1 8.6e+03 0.13 1 0.15 +
2 8.6e+03 0.13 0.5 -0.35 -
3 8.1e+03 0.014 5 0.98 ++
4 8.1e+03 0.014 0.91 -1.3 -
5 8.1e+03 0.021 9.1 0.91 ++
6 8e+03 0.0027 91 1.1 ++
7 8e+03 0.00081 9.1e+02 1 ++
8 8e+03 4.3e-06 9.1e+02 1 ++
Optimization algorithm has converged.
Relative gradient: 4.3017252378782595e-06
Cause of termination: Relative gradient = 4.3e-06 <= 6.1e-06
Number of function evaluations: 24
Number of gradient evaluations: 15
Number of hessian evaluations: 7
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 9
Proportion of Hessian calculation: 7/7 = 100.0%
Optimization time: 0:00:05.560448
Calculate second derivatives and BHHH
File b09post_processing_000010.html has been generated.
File b09post_processing_000010.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000011
*** Initial values of the parameters are obtained from the file __b09post_processing_000011.iter
Cannot read file __b09post_processing_000011.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. Function Relgrad Radius Rho
0 9.1e+03 0.25 1 0.7 +
1 8.5e+03 0.12 1 0.54 +
2 8.2e+03 0.034 1 0.83 +
3 8.1e+03 0.017 1 0.66 +
4 8.1e+03 0.0038 10 1 ++
5 8.1e+03 0.0053 10 0.88 +
6 8.1e+03 5.8e-05 1e+02 1 ++
7 8.1e+03 6.3e-07 1e+02 1 ++
Optimization algorithm has converged.
Relative gradient: 6.345269858865713e-07
Cause of termination: Relative gradient = 6.3e-07 <= 6.1e-06
Number of function evaluations: 25
Number of gradient evaluations: 17
Number of hessian evaluations: 8
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 8
Proportion of Hessian calculation: 8/8 = 100.0%
Optimization time: 0:00:02.639345
Calculate second derivatives and BHHH
File b09post_processing_000011.html has been generated.
File b09post_processing_000011.yaml has been generated.
Biogeme parameters provided by the user.
No yaml file has been found for model b09post_processing_000012
*** Initial values of the parameters are obtained from the file __b09post_processing_000012.iter
Cannot read file __b09post_processing_000012.iter. Statement is ignored.
Starting values for the algorithm: {}
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. Function Relgrad Radius Rho
0 9.2e+03 0.2 1 0.61 +
1 9.2e+03 0.2 0.5 -0.84 -
2 9.1e+03 0.24 0.5 0.11 +
3 8.5e+03 0.099 0.5 0.54 +
4 8.3e+03 0.085 0.5 0.65 +
5 8.1e+03 0.012 5 1 ++
6 8.1e+03 0.012 1.2 -2.8 -
7 8.1e+03 0.022 12 0.93 ++
8 8e+03 0.007 1.2e+02 1.2 ++
9 8e+03 0.0023 1.2e+03 1.1 ++
10 8e+03 0.00012 1.2e+04 1 ++
11 8e+03 1.4e-06 1.2e+04 1 ++
Optimization algorithm has converged.
Relative gradient: 1.3617002739334765e-06
Cause of termination: Relative gradient = 1.4e-06 <= 6.1e-06
Number of function evaluations: 33
Number of gradient evaluations: 21
Number of hessian evaluations: 10
Algorithm: Newton with trust region for simple bound constraints
Number of iterations: 12
Proportion of Hessian calculation: 10/10 = 100.0%
Optimization time: 0:00:08.464501
Calculate second derivatives and BHHH
File b09post_processing_000012.html has been generated.
File b09post_processing_000012.yaml has been generated.
We retrieve the first estimation results for illustration.
spec, results = next(iter(all_results.items()))
print(spec)
asc:GA;b_cost_gen_altspec:altspec;b_time:no_seg;b_time_gen_altspec:generic;model_catalog:nested existing;train_tt_catalog:boxcox
print(results.short_summary())
Results for model b09post_processing_000000
Nbr of parameters: 10
Sample size: 10719
Excluded data: 9
Final log likelihood: -8159.232
Akaike Information Criterion: 16338.46
Bayesian Information Criterion: 16411.26
estimated_parameters = get_pandas_estimated_parameters(estimation_results=results)
display(estimated_parameters)
Name Value ... Robust t-stat. Robust p-value
0 asc_train_ref -0.337820 ... -4.295228 1.745137e-05
1 asc_train_diff_GA 1.049716 ... 11.124844 0.000000e+00
2 b_time -1.548603 ... -26.610823 0.000000e+00
3 lambda_travel_time 0.359844 ... 5.295211 1.188789e-07
4 b_cost_train -0.813968 ... -8.817720 0.000000e+00
5 mu_existing 1.598688 ... 18.413032 0.000000e+00
6 asc_car_ref -0.090481 ... -1.454429 1.458275e-01
7 asc_car_diff_GA -0.502957 ... -4.009956 6.073007e-05
8 b_cost_car -0.339878 ... -4.951261 7.373413e-07
9 b_cost_swissmetro -0.642446 ... -13.224771 0.000000e+00
[10 rows x 5 columns]
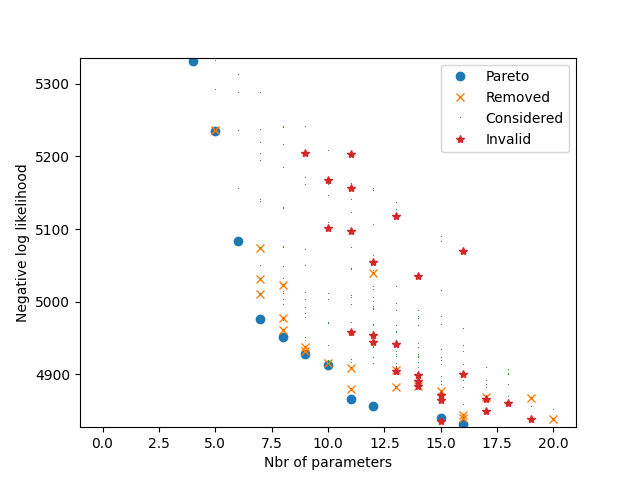
The following plot illustrates all models that have been estimated. Each dot corresponds to a model. The x-coordinate corresponds to the Akaike Information Criterion (AIC). The y-coordinate corresponds to the Bayesian Information Criterion (BIC). Note that there is a third objective that does not appear on this picture: the number of parameters. If the shape of the dot is a circle, it means that it corresponds to a Pareto optimal model. If the shape is a cross, it means that the model has been Pareto optimal at some point during the algorithm and later removed as a new model dominating it has been found. If the shape is a start, it means that the model has been deemed invalid.
if can_plot:
_ = post_processing.plot(
label_x='Nbr of parameters',
label_y='Negative log likelihood',
objective_x=1,
objective_y=0,
)
Total running time of the script: (0 minutes 58.688 seconds)