Note
Go to the end to download the full example code.
Re-estimate the Pareto optimal models¶
The assisted specification algorithm generates a file containing the pareto optimal specification. This script is designed to re-estimate the Pareto optimal models. The catalog of specifications is defined in Specification of a catalog of models .
Michel Bierlaire, EPFL Sat Jun 28 2025, 12:35:57m
from biogeme.results_processing import compile_estimation_results
try:
import matplotlib.pyplot as plt
can_plot = True
except ModuleNotFoundError:
can_plot = False
from biogeme.assisted import ParetoPostProcessing
from plot_b22multiple_models_spec import the_biogeme
PARETO_FILE_NAME = 'saved_results/b22multiple_models.pareto'
CSV_FILE = 'b22process_pareto.csv'
SEP_CSV = ','
The constructor of the Pareto post-processing object takes two arguments:
the biogeme object,
the name of the file where the algorithm has stored the estimated models.
the_pareto_post = ParetoPostProcessing(
biogeme_object=the_biogeme,
pareto_file_name=PARETO_FILE_NAME,
)
the_pareto_post.log_statistics()
all_results = the_pareto_post.reestimate(recycle=True)
summary, description = compile_estimation_results(all_results, use_short_names=True)
print(summary)
Model_000000
Number of estimated parameters 9
Sample size 6768
Final log likelihood -4810.59
Akaike Information Criterion 9639.18
Bayesian Information Criterion 9700.56
asc_train_ref (t-test) -0.0737 (-0.705)
asc_train_diff_male (t-test) -1.16 (-13.7)
asc_train_diff_with_ga (t-test) 2.11 (23.1)
b_time (t-test) -1.61 (-20.2)
b_cost (t-test) -1.49 (-18.4)
b_headway (t-test) -0.00664 (-6.15)
asc_car_ref (t-test) -0.696 (-6.52)
asc_car_diff_male (t-test) 0.474 (4.39)
asc_car_diff_with_ga (t-test) -2 (-9.32)
print(f'Summary table available in {CSV_FILE}')
summary.to_csv(CSV_FILE, sep=SEP_CSV)
Summary table available in b22process_pareto.csv
Explanation of the short names of the models.
with open(CSV_FILE, 'a', encoding='utf-8') as f:
print('\n\n', file=f)
for k, v in description.items():
if k != v:
print(f'{k}: {v}')
print(f'{k}{SEP_CSV}{v}', file=f)
Model_000000: asc:no_seg;train_cost_catalog:linear;train_headway_catalog:without_headway;train_tt_catalog:linear
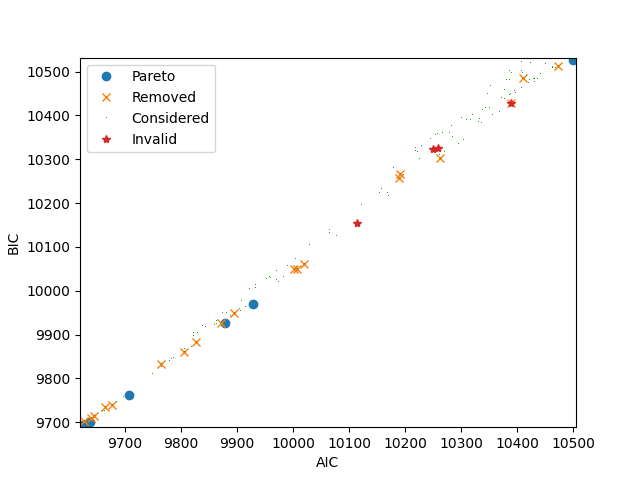
The following plot illustrates all models that have been estimated. Each dot corresponds to a model. The x-coordinate corresponds to the Akaike Information Criterion (AIC). The y-coordinate corresponds to the Bayesian Information Criterion (BIC). Note that there is a third objective that does not appear on this picture: the number of parameters. If the shape of the dot is a circle, it means that it corresponds to a Pareto optimal model. If the shape is a cross, it means that the model has been Pareto optimal at some point during the algorithm and later removed as a new model dominating it has been found. If the shape is a start, it means that the model has been deemed invalid.
if can_plot:
_ = the_pareto_post.plot(label_x='AIC', label_y='BIC')
plt.show()
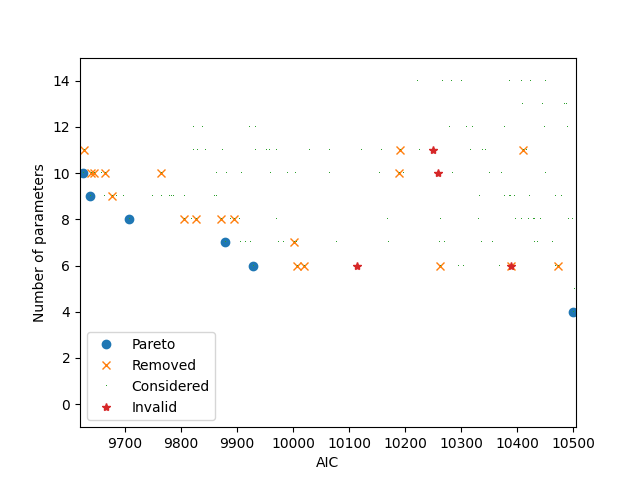
It is possible to plot two different objectives: AIC and number of parameters.
if can_plot:
_ = the_pareto_post.plot(
objective_x=0, objective_y=2, label_x='AIC', label_y='Number of parameters'
)
plt.show()
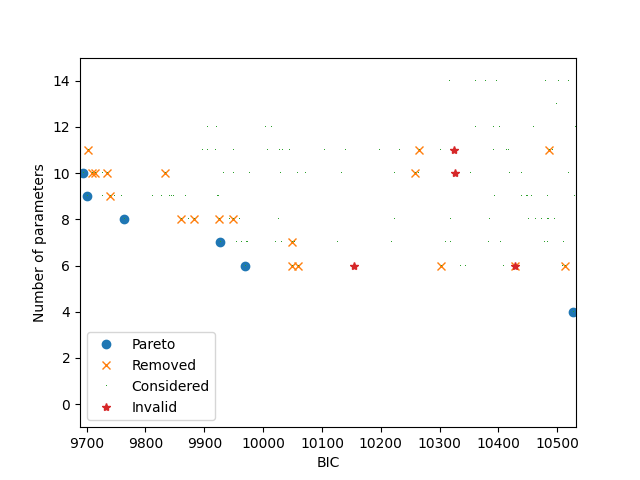
It is possible to plot two different objectives: BIC and number of parameters.
if can_plot:
_ = the_pareto_post.plot(
objective_x=1, objective_y=2, label_x='BIC', label_y='Number of parameters'
)
plt.show()
Total running time of the script: (0 minutes 0.248 seconds)