Note
Go to the end to download the full example code.
Re-estimate the Pareto optimal models
The assisted specification algorithm generates a file containing the pareto optimal specification. This script is designed to re-estimate the Pareto optimal models. The catalog of specifications is defined in Specification of a catalog of models .
- author:
Michel Bierlaire, EPFL
- date:
Wed Apr 12 17:46:14 2023
import biogeme.biogeme_logging as blog
try:
import matplotlib.pyplot as plt
can_plot = True
except ModuleNotFoundError:
can_plot = False
from biogeme_optimization.exceptions import OptimizationError
from biogeme.assisted import ParetoPostProcessing
from biogeme.results import compile_estimation_results
from plot_b21multiple_models_spec import the_biogeme
PARETO_FILE_NAME = 'saved_results/b21multiple_models.pareto'
logger = blog.get_screen_logger(blog.INFO)
logger.info('Example b21process_pareto.py')
CSV_FILE = 'b21process_pareto.csv'
SEP_CSV = ','
Example b21process_pareto.py
The constructor of the Pareto post processing object takes two arguments:
the biogeme object,
the name of the file where the algorithm has stored the estimated models.
the_pareto_post = ParetoPostProcessing(
biogeme_object=the_biogeme,
pareto_file_name=PARETO_FILE_NAME,
)
Pareto set initialized from file with 36 elements [8 Pareto] and 0 invalid elements.
the_pareto_post.log_statistics()
Pareto: 8
Considered: 36
Removed: 4
Complete re-estimation of the best models, including the calculation of the statistics.
all_results = the_pareto_post.reestimate(recycle=False)
Biogeme parameters provided by the user.
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
*** Initial values of the parameters are obtained from the file __b21multiple_models_000000.iter
Parameter values restored from __b21multiple_models_000000.iter
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. ASC_CAR ASC_CAR_GA ASC_TRAIN ASC_TRAIN_GA B_COST B_TIME lambda_time Function Relgrad Radius Rho
0 -0.062 -0.34 -0.94 1.9 -1.1 -1.7 0.36 5e+03 0.0063 10 1 ++
1 -0.064 -0.31 -1 2 -1.1 -1.7 0.38 5e+03 0.00022 1e+02 1 ++
2 -0.064 -0.31 -1 2 -1.1 -1.7 0.38 5e+03 2.2e-07 1e+02 1 ++
Results saved in file b21multiple_models_000000~00.html
Results saved in file b21multiple_models_000000~00.pickle
Biogeme parameters provided by the user.
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
*** Initial values of the parameters are obtained from the file __b21multiple_models_000001.iter
Parameter values restored from __b21multiple_models_000001.iter
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. ASC_CAR ASC_CAR_GA ASC_CAR_male ASC_TRAIN ASC_TRAIN_GA ASC_TRAIN_male B_COST B_TIME lambda_time Function Relgrad Radius Rho
0 -0.42 -0.45 0.41 -0.22 2 -1.1 -1.1 -1.7 0.34 4.9e+03 0.0002 10 1 ++
1 -0.42 -0.45 0.41 -0.22 2 -1.1 -1.1 -1.7 0.34 4.9e+03 1.9e-07 10 1 ++
Results saved in file b21multiple_models_000001~00.html
Results saved in file b21multiple_models_000001~00.pickle
Biogeme parameters provided by the user.
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
*** Initial values of the parameters are obtained from the file __b21multiple_models_000002.iter
Parameter values restored from __b21multiple_models_000002.iter
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Results saved in file b21multiple_models_000002~00.html
Results saved in file b21multiple_models_000002~00.pickle
Biogeme parameters provided by the user.
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
*** Initial values of the parameters are obtained from the file __b21multiple_models_000003.iter
Parameter values restored from __b21multiple_models_000003.iter
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. ASC_CAR ASC_CAR_GA ASC_CAR_male ASC_TRAIN ASC_TRAIN_GA ASC_TRAIN_male B_COST B_COST_GA B_TIME lambda_time Function Relgrad Radius Rho
0 -0.4 -0.8 0.39 -0.25 1.9 -1.1 -1 0.89 -1.6 0.39 4.9e+03 0.012 10 1 ++
1 -0.42 -1 0.41 -0.22 2 -1.2 -1.1 0.92 -1.7 0.33 4.9e+03 0.0004 1e+02 1 ++
2 -0.42 -1 0.41 -0.22 2 -1.2 -1.1 0.92 -1.7 0.33 4.9e+03 2.5e-06 1e+02 1 ++
Results saved in file b21multiple_models_000003~00.html
Results saved in file b21multiple_models_000003~00.pickle
Biogeme parameters provided by the user.
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
*** Initial values of the parameters are obtained from the file __b21multiple_models_000004.iter
Parameter values restored from __b21multiple_models_000004.iter
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. ASC_CAR ASC_CAR_GA ASC_CAR_male ASC_TRAIN ASC_TRAIN_GA ASC_TRAIN_male B_COST B_COST_inc-100+ B_COST_inc-50-1 B_COST_inc-unde B_COST_inc-unkn B_TIME lambda_time Function Relgrad Radius Rho
0 -0.46 -0.32 0.45 -0.28 2 -1.1 -1.5 0.58 0.2 -0.62 0.79 -1.7 0.33 4.9e+03 0.0021 10 1 ++
1 -0.46 -0.32 0.45 -0.28 2 -1.1 -1.5 0.58 0.2 -0.62 0.79 -1.7 0.33 4.9e+03 3.5e-05 10 1 ++
Results saved in file b21multiple_models_000004~00.html
Results saved in file b21multiple_models_000004~00.pickle
Biogeme parameters provided by the user.
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
*** Initial values of the parameters are obtained from the file __b21multiple_models_000005.iter
Parameter values restored from __b21multiple_models_000005.iter
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. ASC_CAR ASC_TRAIN B_COST B_TIME Function Relgrad Radius Rho
0 -0.072 -0.73 -0.93 -1.2 5.3e+03 0.017 1 0.82 +
1 -0.15 -0.71 -1.1 -1.3 5.3e+03 0.0009 10 1 ++
2 -0.15 -0.71 -1.1 -1.3 5.3e+03 3.9e-06 10 1 ++
Results saved in file b21multiple_models_000005~00.html
Results saved in file b21multiple_models_000005~00.pickle
Biogeme parameters provided by the user.
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
*** Initial values of the parameters are obtained from the file __b21multiple_models_000006.iter
Parameter values restored from __b21multiple_models_000006.iter
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. ASC_CAR ASC_TRAIN B_COST B_TIME lambda_time Function Relgrad Radius Rho
0 -0.0036 -0.37 -1.1 -1.7 0.5 5.3e+03 0.016 1 0.83 +
1 -0.0049 -0.48 -1.1 -1.7 0.51 5.3e+03 0.00057 10 1 ++
2 -0.0049 -0.48 -1.1 -1.7 0.51 5.3e+03 8.2e-07 10 1 ++
Results saved in file b21multiple_models_000006~00.html
Results saved in file b21multiple_models_000006~00.pickle
Biogeme parameters provided by the user.
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
*** Initial values of the parameters are obtained from the file __b21multiple_models_000007.iter
Parameter values restored from __b21multiple_models_000007.iter
As the model is not too complex, we activate the calculation of second derivatives. If you want to change it, change the name of the algorithm in the TOML file from "automatic" to "simple_bounds"
Optimization algorithm: hybrid Newton/BFGS with simple bounds [simple_bounds]
** Optimization: Newton with trust region for simple bounds
Iter. ASC_CAR ASC_CAR_GA ASC_CAR_male ASC_TRAIN ASC_TRAIN_GA ASC_TRAIN_male B_COST B_TIME Function Relgrad Radius Rho
0 -0.42 -0.45 0.41 -0.22 2 -1.2 -1.1 -1.7 4.9e+03 3.7e-05 1 1
Results saved in file b21multiple_models_000007~00.html
Results saved in file b21multiple_models_000007~00.pickle
summary, description = compile_estimation_results(all_results, use_short_names=True)
print(summary)
Model_000000 ... Model_000007
Number of estimated parameters 7 ... 8
Sample size 6768 ... 6768
Final log likelihood -4995.755387 ... -4900.883444
Akaike Information Criterion 10005.510775 ... 9817.766888
Bayesian Information Criterion 10053.250501 ... 9872.326575
ASC_CAR (t-test) -0.064 (-1.22) ... -0.389 (-3.95)
ASC_CAR_GA (t-test) -0.313 (-1.59) ... -0.415 (-2.02)
ASC_TRAIN (t-test) -1.03 (-13.9) ... -0.203 (-2.23)
ASC_TRAIN_GA (t-test) 2.04 (22.8) ... 2.03 (22.4)
B_COST (t-test) -1.1 (-14.8) ... -1.06 (-15.2)
B_TIME (t-test) -1.67 (-21.3) ... -1.7 (-21.5)
lambda_time (t-test) 0.382 (5.18) ...
ASC_CAR_male (t-test) ... 0.377 (3.65)
ASC_TRAIN_male (t-test) ... -1.2 (-14.1)
B_COST_GA (t-test) ...
B_COST_inc-100+ (t-test) ...
B_COST_inc-50-100 (t-test) ...
B_COST_inc-under50 (t-test) ...
B_COST_inc-unknown (t-test) ...
[19 rows x 8 columns]
print(f'Summary table available in {CSV_FILE}')
summary.to_csv(CSV_FILE, sep=SEP_CSV)
Summary table available in b21process_pareto.csv
Explanation of the short names of the models.
with open(CSV_FILE, 'a', encoding='utf-8') as f:
print('\n\n', file=f)
for k, v in description.items():
if k != v:
print(f'{k}: {v}')
print(f'{k}{SEP_CSV}{v}', file=f)
Model_000000: ASC:GA;B_COST:no_seg;TRAIN_TT:boxcox
Model_000001: ASC:MALE-GA;B_COST:no_seg;TRAIN_TT:boxcox
Model_000002: ASC:GA;B_COST:no_seg;TRAIN_TT:log
Model_000003: ASC:MALE-GA;B_COST:GA;TRAIN_TT:boxcox
Model_000004: ASC:MALE-GA;B_COST:INCOME;TRAIN_TT:boxcox
Model_000005: ASC:no_seg;B_COST:no_seg;TRAIN_TT:linear
Model_000006: ASC:no_seg;B_COST:no_seg;TRAIN_TT:boxcox
Model_000007: ASC:MALE-GA;B_COST:no_seg;TRAIN_TT:log
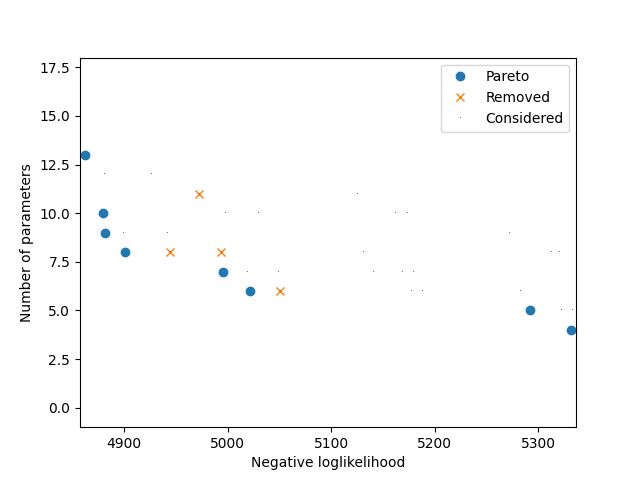
The following plot illustrates all models that have been estimated. Each dot corresponds to a model. The x-coordinate corresponds to the negative log-likelihood. The y-coordinate corresponds to the number of parameters. If the shape of the dot is a circle, it means that it corresponds to a Pareto optimal model. If the shape is a cross, it means that the model has been Pareto optimal at some point during the algorithm and later removed as a new model dominating it has been found.
if can_plot:
try:
_ = the_pareto_post.plot(
label_x='Negative loglikelihood', label_y='Number of parameters'
)
plt.show()
except OptimizationError as e:
print(f'No plot available: {e}')
Total running time of the script: (0 minutes 1.547 seconds)